Section 6 Cross-disease comparisons
6.1 Prioritisation map
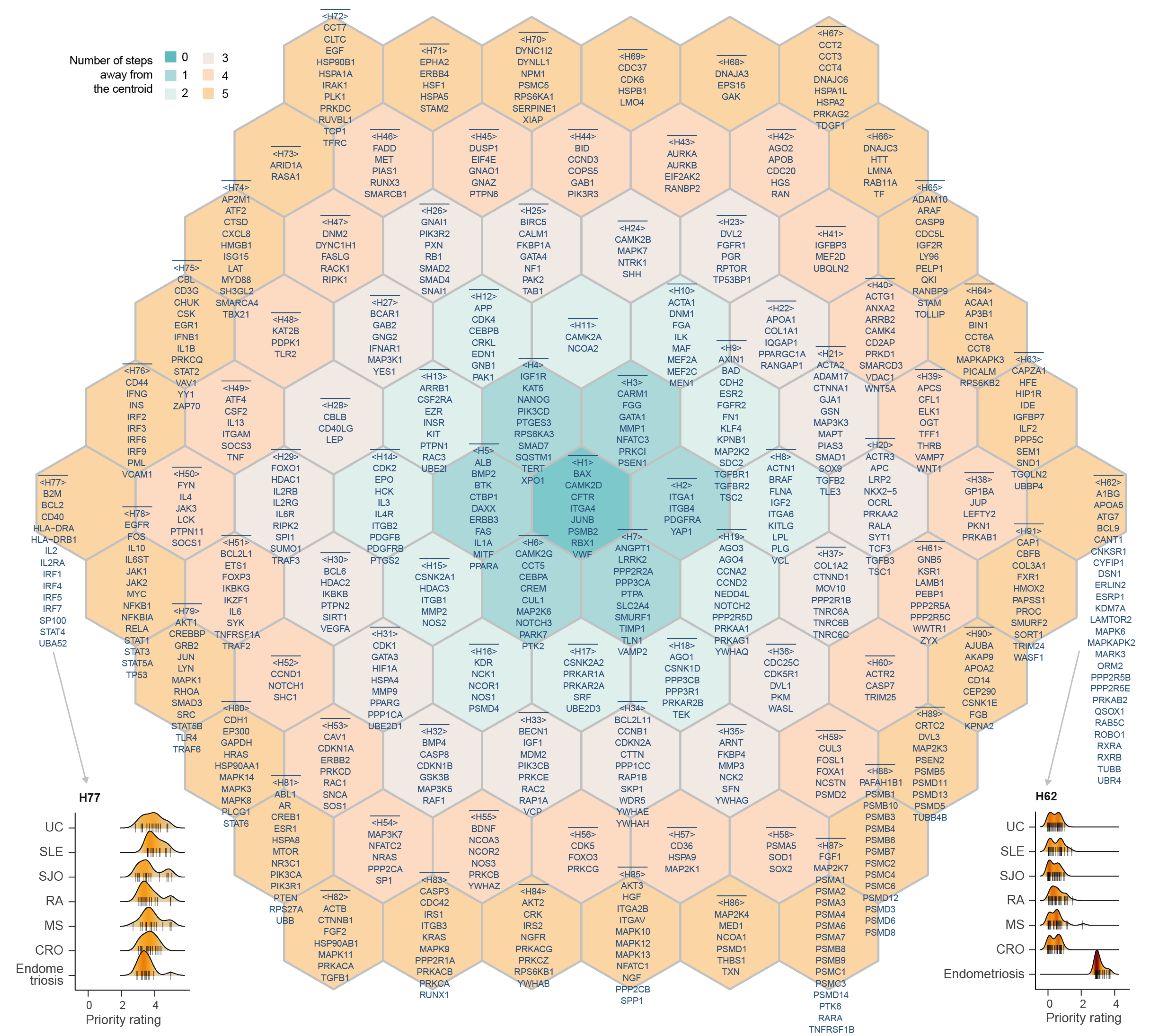
Figure 6.1: Illustration of the supra-hexagonal map, consisting of 91 hexagons indexed circularly outward (H1-H91; such circular indexing is indicated in colors). Also displayed are genes in each hexagon. Beneath are ridge plots for two hexagons, showing density of priority rating for genes in H77 (bottom-left) and in H62 (bottom-right). CRO: Crohn’s Disease; MS: Multiple Sclerosis; RA: Rheumatoid Arthritis; SLE: Systemic Lupus Erythematosus; SJO: Sjögren’s syndrome; UC: Ulcerative Colitis.
6.2 Target clusters
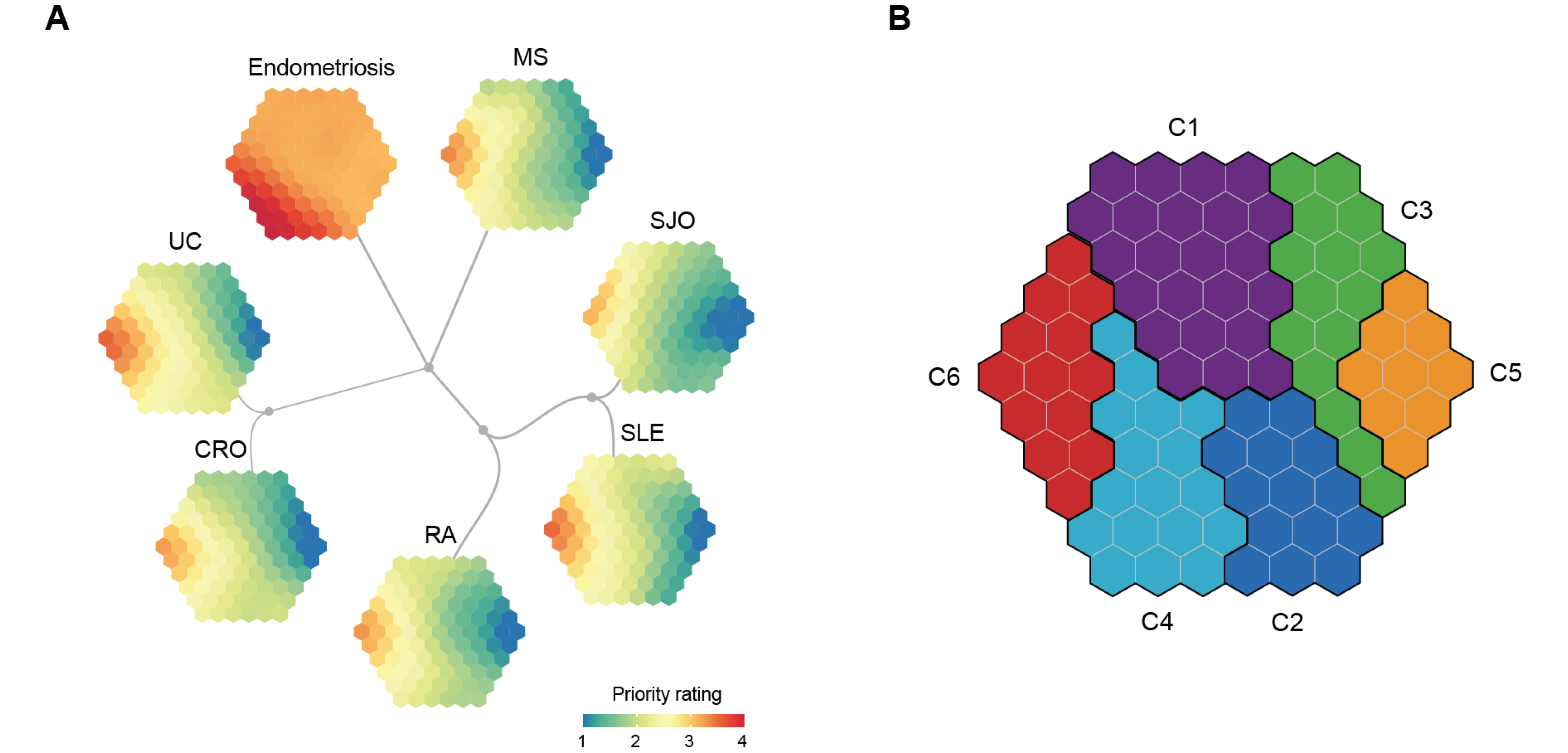
Figure 6.2: Identification of shared and distinct target gene clusters. (A) Each map illustrates illustrating a disease-specific gene prioritisation profile, while consensus neighbour-joining tree captures the similarity of inter-disease prioritisation profiles. (B) Target gene clusters. The prioritisation map divided into 6 clusters (C1-C6), each covering continuous hexagons as color-coded.
6.3 Cluster C6
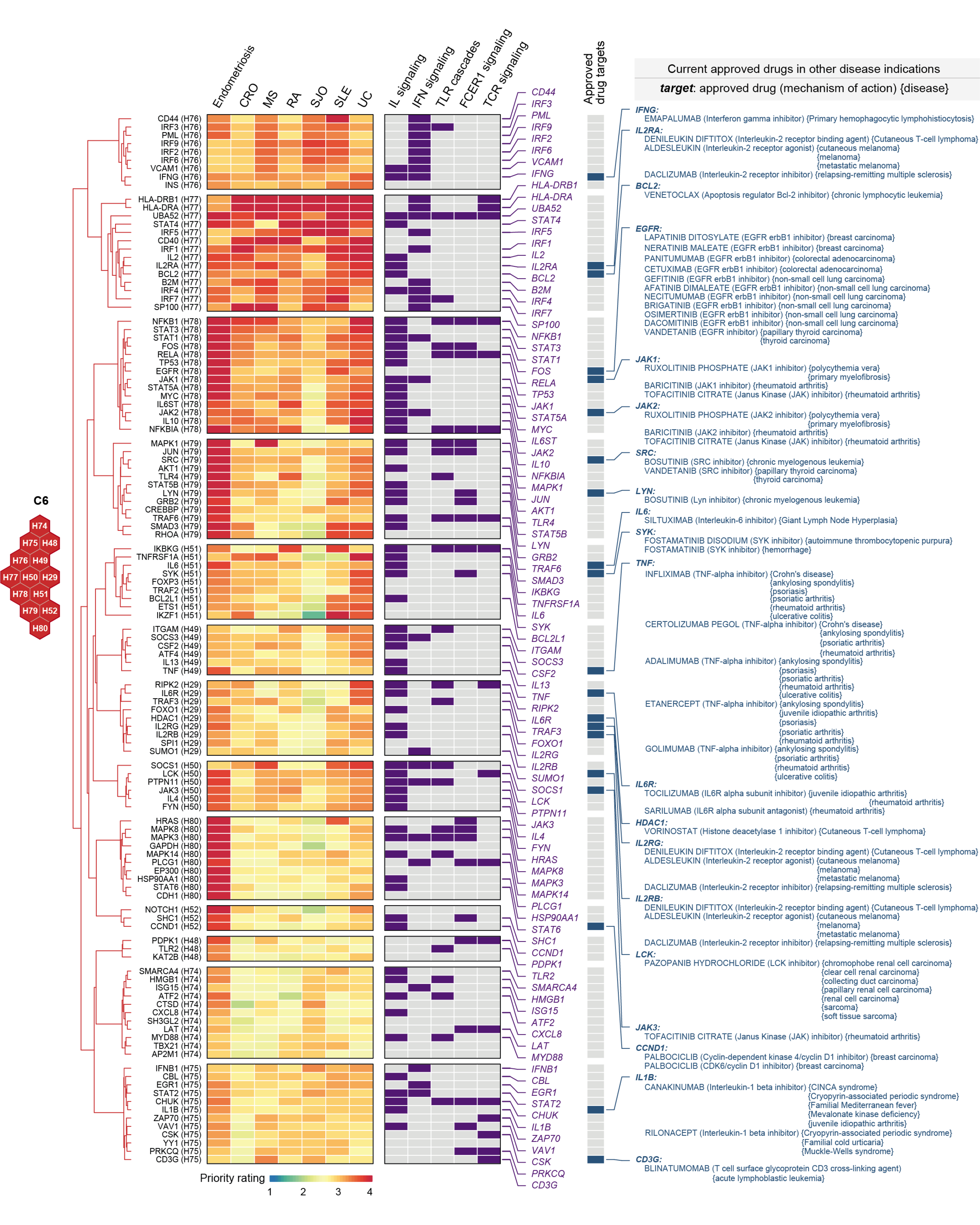
Figure 6.3: Repurposing evidence for immunomodulatory drugs based on shared target genes in the cluster C6. Heatmap illustrates target genes in C6 prioritised across 7 diseases, with annotations to IL, IFN, TLR, FCER1 and TCR signaling pathways, together with information on approved drug targets, repurposed drugs, mechanisms of action and disease indications.
6.4 Cluster C5
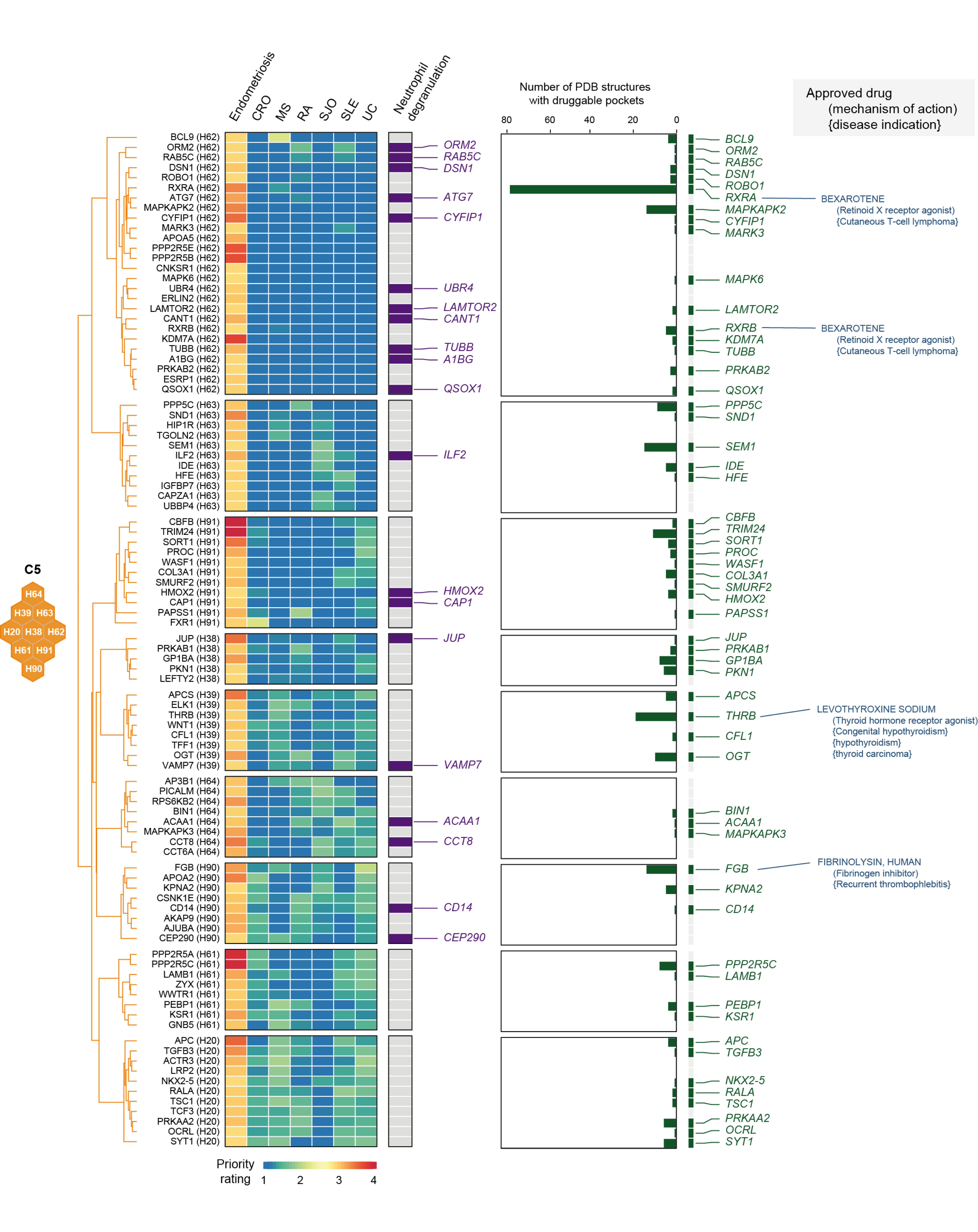
Figure 6.4: Tractable evidence for targeting neutrophil degranulation based on genes prioritised specific to endometriosis in the cluster C5. Heatmap illustrates target genes in C5 prioritised across 7 diseases, together with information on functional relevance to neutrophil degranulation, the tractability (the number of druggable pockets), and therapeutic approval (approved drug targets, repurposed drugs, mechanisms of action and disease indications).
6.5 Clustered genes
Notably, the column END refer to the priority rating for endometriosis.